Key points
- CDC funds 50 states, several cities, and two territory public health labs to perform testing on bacterial and fungal pathogens.
- All seven regional Antimicrobial Resistance Laboratory Network (AR Lab Network) labs perform a core set of standard tests.
- To use AR Lab Network services, contact your local or state healthcare-associated infection (HAI) coordinator, or AR regional lab.

Testings offered
Core testing performed by seven regional labs
- Molecular testing to detect colonization from carbapenemase-producing organisms (CPO) or C. auris to prevent spread.
- Identification and antifungal susceptibility testing (AFST) of Candida species to identify emerging resistance.
- Identification, antibiotic susceptibility testing (AST), mechanism testing, and carbapenemase-production testing of CPOs to identify emerging resistance.
- Whole genome sequence (WGS) testing of healthcare-associated Gram-negative bacteria and C. auris to identify new resistance and understand spread.
- Detection and characterization of emerging and concerning threats.
- Expanded susceptibility testing using new and/or new combination of antimicrobials against metallo-β-lactamase (MBL)-producing Enterobacterales and multidrug-resistant fungi.
Additional testing performed by select regional labs
- Special projects for C. difficile.
- Antimicrobial susceptibility testing (AST), WGS and select molecular testing of drug-resistant N. gonorrhoeae.
- AST and serotyping of multidrug-resistant S. pneumoniae.
- WGS of all U.S. M. tuberculosis isolates.
- Screening A. fumigatus isolates for azole resistance, with confirmatory testing performed on isolates that screen positive.
- Organism ID, AFST, and WGS for dermatophytes including Trichophyton, Epidermophyton, and Microsporum.
Descriptions of testing
Colonization screening
- Some people can carry pathogens without developing an infection or showing symptoms, known as colonization; people who are colonized with the pathogens can spread them to others without knowing.
- When unusual resistance is identified in an individual, healthcare workers use lab tests to screen other patients to see if they are colonized with the same antimicrobial-resistant pathogen, which can prompt additional infection control actions.
Identification of pathogens
- Labs identify and confirm the genus and species of resistant pathogens.
Molecular testing
- A collection of techniques used to detect specific genes within a pathogen, including those that have and can share resistance.
Carbapenemase activity detection test
- This is a phenotypic test used to assess whether carbapenemase enzyme's activity is present in an organism. Many bacteria produce this enzyme, not just bacterial pathogens.
- If a carbapenemase is produced, then a carbapenem antibiotic will not kill the pathogen.
Antimicrobial susceptibility testing (AST) and antifungal susceptibility testing (AFST)
- This is a type of lab test that determines how sensitive a pathogen is to different antibiotics or antifungals.
- These tests can help select the best drug for treating an antimicrobial-resistant infection and provide data to monitor how a pathogen's resistance to that drug (and others) might change over time.
Whole genome sequencing (WGS)
- A laboratory procedure that provides a very precise DNA fingerprint to help link cases to one another, helping to identify or confirm transmission.
- WGS allows scientists to understand the specific type of resistance mechanisms present and understand how common or uncommon specific DNA fingerprints are.
Carbapenemase-producing Organisms (CPO)
CPOs are types of bacteria that produce a genetically coded enzyme called carbapenemase. Carbapenemases allow these bacteria to resist the effects of carbapenems and other β-lactam antibiotics. Carbapenems are among the few remaining antibiotics that can treat extended-spectrum β-lactamase (ESBL)-producing bacteria, but resistance is on the rise. Detecting patients who are colonized with CPOs or have an infection caused by a CPO helps prevent spread within and among facilities.
CPOs screened:
Screening for CPO colonization
Who performs the test
- Some local or state public health labs.
- The seven AR Lab Network regional labs.
About this testing
- Screening test is free, including shipping.
- High-risk patients include those who have had contact with another patient diagnosed with a CPO or other patient contacts considered to be high risk (e.g., received health care in a foreign country in the past six months).
- CPO colonization screening is performed using rectal swabs (FDA-approved Cepheid GeneXpert Carba-R test kit) to detect blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP-1 genes.
- Culture-based screening can be used to detect additional genes or variants (i.e., other IMP variants or other OXA genes circulating in the U.S.).
Request testing
- Public health officials, healthcare facilities, infection preventionists, and epidemiologists can request colonization screening of high-risk patients through their HAI coordinator.
- Screening results are returned to the submitting facility, jurisdictional health department, and state public health lab within two business days of specimen receipt for PCR screening.
Testing CRO isolates
Who performs the tests
- An estimated 56 local or state public health labs.
- The seven AR Lab Network regional labs.
About this testing
- Testing is free, including shipping.
- For Enterobacterales isolates that are resistant to ertapenem, imipenem, doripenem, or meropenem; and Pseudomonas aeruginosa isolates that are resistant to imipenem, doripenem, or meropenem, using standard susceptibility testing methods (i.e., minimal inhibitory concentrations of ≥8 µg/mL), the following is performed:
- Organism confirmation (e.g., MALDI-TOF mass spectrometry).
- Carbapenemase production using the mCIM or CarbaNP test.
- Real time PCR (RT-PCR) to detect targeted carbapenemase genes (KPC, NDM, VIM, OXA-48, and IMP).
- Antimicrobial susceptibility testing using a broad panel of drugs by broth microdilution, Kirby-Bauer testing, and/or gradient diffusion strips.
Request testing
- Healthcare providers, and clinical labs can contact their state or local public health lab to request testing.
- Results of the isolate test are reported to the submitting lab or facility.
- Public health labs will immediately notify public health officials with concerning novel or emerging resistance is detected (i.e., pan-resistance, rare, or potentially novel carbapenemase genes.)
Azole-resistant Aspergillus fumigatus (A. fumigatus)
A. fumigatus, a fungus found in the environment, can cause serious illness in people with weakened immune systems. This fungus can develop resistance to azoles during long-term treatment of infected patients or when it is exposed to azole fungicides used in the environment. Infections have been identified in the U.S., increasing the need for awareness and action by clinical and public health professionals. The AR Lab Network tests for azole resistance in A. fumigatus in clinical isolates to monitor and track the emergence of azole resistance in the U.S.
Testing for A. fumigatus
Who performs the tests
- CDC's AR Lab Network regional labs at the Maryland Public Health Laboratory and the Tennessee State Public Health Laboratory perform testing nationwide for azole resistance in A. fumigatus.
About this testing
- Testing is free, including shipping.
- Testing includes:
- Species confirmation.
- Screening for azole resistance using an agar plating method.
- Confirmatory testing via broth microdilution for the following antifungals: posaconazole, voriconazole, itraconazole, and isavuconazole.
- Healthcare and clinical labs should submit:
- Clinical A. fumigatus isolates from any body site, especially if there was concern for an invasive or azole-resistant infection.
- If AFST was done previously, all A. fumigatus isolates with AFST results indicating azole resistance from any body site (invasive or non-invasive, sterile or non-sterile).
- Only submit A. fumigatus or suspected A. fumigatus isolates; do not submit isolates identified as unspecified Aspergillus species or as another Aspergillus species.
Request testing
- Healthcare, clinical, reference, and public health labs can contact your state public health laboratory, the Maryland Public Health Laboratory, or the Tennessee State Public Health Laboratory to request testing.
- State requirements vary regarding submitting isolates to the state public health lab or directly to an AR Lab Network Aspergillus Laboratory.
- Results will be reported back to the submitting institution within two weeks of receipt of the isolate.
Candida, including Candida auris
Candida is one of the most common causes of healthcare-associated bloodstream infections in the U.S., and antifungal resistance in Candida is increasing. New and emerging species, like Candida auris (C. auris), can spread in healthcare settings and cause outbreaks.
Testing Candida isolates
Who performs the test
- Some local or state public health labs.
- The seven AR Lab Network regional labs.
About this testing
- Testing is free, including shipping.
- Healthcare, clinical, and public health laboratories can submit the following isolates:
- Suspected C. auris from any specimen site.
- Candida species other than Candida albicans from any specimen site, particularly invasive sites.
- Yeast isolates that could not be identified by the submitting laboratory.
- AR Lab Network regional labs:
- Confirm Candida species using MALDI-TOF mass spectrometry or DNA sequencing.
- Perform antifungal susceptibility testing using a CDC-developed, custom broth microdilution panel.
- Some isolates may also be tested against new antifungals, known as expanded AFST, to help understand if a new drug can effectively treat highly-resistant infections.
- Report confirmed C. auris cases immediately to public health authorities for containment response.
- Provide whole genome sequencing to track and characterize resistance and strengthen epidemiology investigations to contain outbreaks.
- Not all local or state public health labs can perform the same isolate testing as AR Lab Network regional labs. Therefore, isolates sent to local or state public health labs may receive some testing before being sent to a regional lab for further testing.
Request testing
- To request testing healthcare, clinical, and public health lab facilities should contact their state public health lab or AR Lab Network regional lab.
Screening for C. auris colonization
Who performs the test
- Some local or state public health labs.
- The seven AR Lab Network regional labs.
About this testing
- This screening is free of charge, including shipping.
- Healthcare facilities can request C. auris colonization screening of close contacts of an index patient (first patient identified with an infection) or other patient contacts considered high-risk (e.g., those requiring mechanical ventilation).
- High-risk patients include those who have had contact with another patient diagnosed with C. auris or other patient contacts considered to be high risk (e.g., received healthcare in a foreign country in the past six months).
- C. auris colonization screening is performed using a swab specimen, typically a combined bilateral axillary-groin swab.
Request testing
- Public health officials (e.g., state HAI coordinators), healthcare facilities, infection preventionists, and epidemiologists can request colonization screening of high-risk patients. To request screening and order screening materials, contact your state-based healthcare-associated infection program coordinator.
- Screening results are returned to submitting institution within two business days of specimen receipt for PCR screening.
C. difficile
C. difficile causes severe diarrhea and colitis (an inflammation of the colon), which can be life-threatening. Most cases of infection occur in people taking antibiotics or those who have recently finished taking antibiotics.
Testing for C. difficile
Who performs the test
- CDC's AR Lab Network regional labs at the Minnesota Department of Health Public Health Laboratory and the Wadsworth Center Laboratories can help with outbreak investigations or special projects related to C. difficile.
About this testing
- CDC's AR Lab Network's C. difficile labs can provide outbreak investigation support by performing C. difficile isolation, culturing, and whole genome sequencing to assess transmission.
Request testing
- Healthcare facilities can contact the AR Lab Network regional labs at the Minnesota Department of Health Public Health Laboratory and the Wadsworth Center Laboratories.
Expanded AST for Hard-to-Treat Infections
When antibiotics are newly approved for use, it can take years before susceptibility tests for these antibiotics are made available in clinical labs. CDC's AR Lab Network provides testing for isolates of metallo-beta-lactamase (MBL) producing-Enterobacterales, which can help guide clinical decision making for highly-resistant isolates. MBLs make bacteria resistant to a broad range of antibiotics, including carbapenems, which are often used to treat antimicrobial-resistant infections.
Testing for MBL isolates
Who performs the test
- The seven AR Lab Network regional labs.
About this testing
- This testing is free.
- Using the Hewlett-Packard D300e Digital Dispenser, highly-resistant Enterobacterales isolates will be tested for susceptibility to the combination of aztreonam and avibactam.
- The combination drug aztreonam/avibactam is still in clinical development. However, because of limited treatment options, the IDSA's Guidance on Treatment of Antimicrobial Resistant Gram-Negative Infections recommends this combination as a treatment option for serious infections caused by MBL-producing CRE.
- Labs can request testing of Enterobacterales isolates that:
- Are not susceptible to all beta-lactams tested, including either ceftazidime/avibactam or meropenem/vaborbactam. These may be MBL-producing isolates that cause infections with few effective treatment options. OR
- Enterobacterales that test positive for NDM, VIM, or IMP genes (blaNDM, blaVIM, or blaIMP) confirmed by a molecular test.
- Requests must be accompanied by preliminary lab testing results, including AST results and/or molecular results, to confirm inclusion criteria have been met.
- Susceptibility results will be reported for ceftazidime/avibactam, aztreonam, and aztreonam/avibactam to help assess utility of combination therapy.
- Isolates will also be tested for:
- Susceptibility to more than 20 other antibiotics using a commercially available comprehensive broth microdilution panel.
- Carbapenemase production using the mCIM.
- MBL and other carbapenemase genes.
Request testing
- Healthcare providers, and clinical and public health labs can contact their AR Lab Network regional lab to request ExAST and learn about alternative treatment options.
- Susceptibility results for confirmed MBL-producing isolates will be returned within three business days.
- The AR Lab Network regional labs will notify the submitter and public health officials if MBL-producing isolates are confirmed to have an aztreonam/avibactam MIC ≥ 8/4 µg/mL.
N. gonorrhoeae (Gonorrhea)
Gonorrhea has progressively developed antimicrobial resistance to the drugs recommended to treat it. CDC's AR Lab Network offers testing to assist in care of patients with potentially drug-resistant gonorrhea.
Testing for N. gonorrhoeae
Who performs the test
- Some local or state public health labs perform testing for N. gonorrhoeae specimens or isolates suspected to be resistant.
- CDC's AR Lab Network regional labs at the Maryland Public Health Laboratory and Washington Public Health Laboratory support labs nationwide, if the local or state lab does not perform this test.
About this test
- Testing is free, including shipping.
- Includes gradient strip AST for four antibiotics: azithromycin, ceftriaxone, cefixime and ciprofloxacin.
Request testing
- Contact your local or state lab to see if this testing is available. If testing is not available, please reach out to your AR Lab Network regional lab.
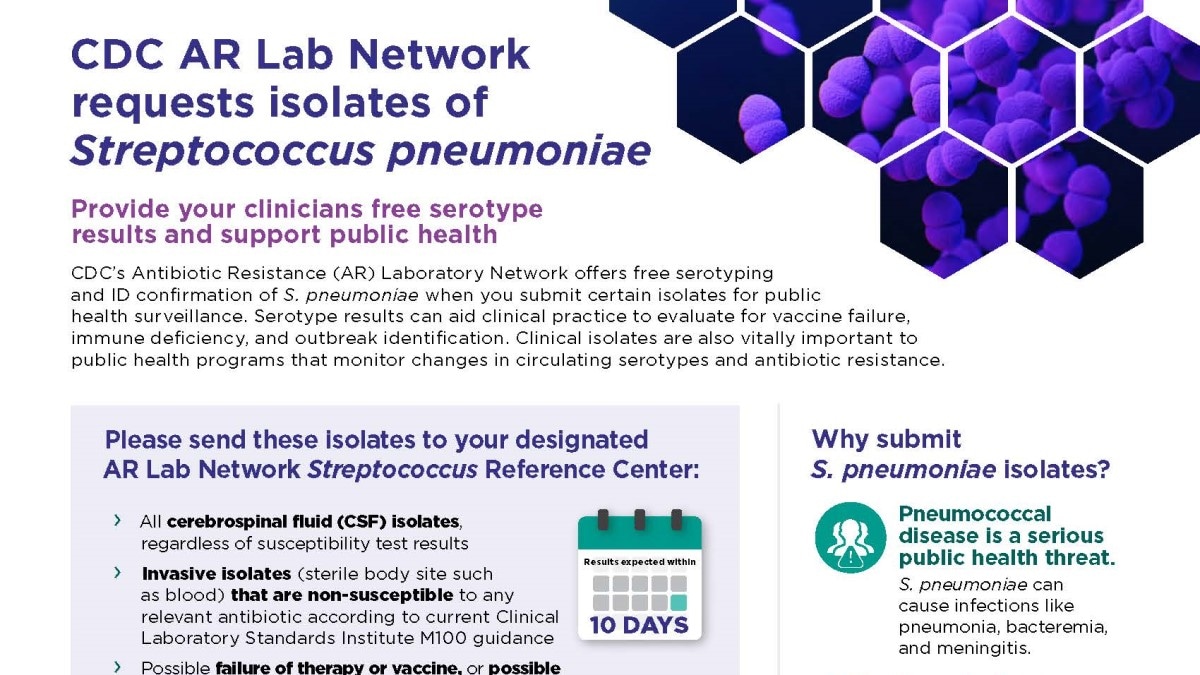
Streptococcus pneumoniae (S. pneumoniae)
S. pneumoniae (pneumococcus) is a leading cause of bacterial pneumonia and meningitis in the U.S. When labs submit pneumococcus isolates for testing, serotype results can aid clinical practice to evaluate for vaccine failure, immune deficiency, and outbreak identification. Clinical isolates are also vitally important to public health programs that monitor changes in circulating serotypes and antimicrobial resistance.
Testing for S. pneumoniae
Who performs the tests
- CDC's AR Lab Network regional labs at the Wisconsin State Laboratory of Hygiene and the Minnesota Department of Health Public Health Laboratory serve as AR Lab Network Streptococcus Reference Centers and perform serotyping and ID confirmation nationwide.
About this testing
- Testing is free, including shipping.
- Testing includes:
- Species identification by phenotypic or molecular methods.
- Serotyping/serogrouping using molecular methods such as conventional and/or real-time PCR or WGS.
- AST by broth microdilution.
- Identification and serotyping results are sent within ten days of specimen receipt. AST results are for surveillance purposes only and AR Labs will not send these results back.
Request testing
- Healthcare and clinical labs can contact their state public health lab, the Wisconsin State Laboratory of Hygiene or the Minnesota Department of Health Public Health Laboratory.
- Healthcare and clinical labs should submit:
- All CSF isolates, regardless of susceptibility test results.
- Invasive isolates (sterile body site such as blood) that are non-susceptible for any clinically relevant antibiotic according to current CLSI M100 guidance.
- Possible failure of therapy or vaccine, or possible outbreak-related isolates.
Testing Mycobacterium tuberculosis (M. tuberculosis)
M. tuberculosis, most commonly known as Tuberculosis (TB), is a disease caused by bacteria that are spread from person to person through the air. It is a common infectious disease found worldwide and can cause death if not treated. TB can be resistant to more than one antibiotic used to treat it.
The National TB Molecular Surveillance Center provides testing to identify TB strains, free of charge.
Request testing
Any authorized medical or laboratory professionals can submit samples, in consultation with their state health department or local authorities and per institutional policies. For required submission forms, visit the TB laboratory information page linked below.
Data Modernization Initiative
CDC is committed to data timeliness and is improving the speed and efficiency of antimicrobial resistance data electronically transmitted from AR Lab Network laboratories to CDC through the Data for Action on Antimicrobial-Resistant Threats (DAART) portal. CDC is also improving the format and usability of data shared with antimicrobial resistance pathogen subject matter experts.
DAART helps experts get antimicrobial resistance data faster to support rapid responses to stop spread and protect people.
Resources for U.S. labs
CDC offers free resources and tools for laboratory scientists to help combat antimicrobial resistance. Email CDC at ARLN@cdc.gov for more information about the AR Lab Network.